How’s it going everyone?! Well, I decided to join Thomas Mock’s #TidyTuesday on twitter and this is my second submission. I’ll write a second post on my first submission because that took a little longer to figure out. Anyway let’s get going! First, let’s import the necessary packages
library(tidyverse)
library(rjson)
colors <- fromJSON(file = "~/Code/tidy_tuesday/Week_3/colors.json")Rjson is only needed for my color palette, which I created as a JSON from here. I also imported my color palette as the vector named colors. Next, since I want to be able to produce as many as these plots as I want, I created a function that would let me avoid redundance. The function first checks whether we’re computing for the entire world or just a country, then does the necessary tidying up and plotting. Of course, in order to create the function I first explored the dataset. This is just the finalized product.
plotting_function <-
function(
data,
yearOfInterest,
countryName,
title = paste("Share of death by cause, ", countryName, ", ", yearOfInterest, sep = ""),
subtitle = "",
caption = "Source: IHME, Global Burden of Disease"
){
if (countryName == "World") {
data <- data %>%
filter(year == yearOfInterest) %>%
select(-c(country, country_code, year)) %>%
summarize_all(funs("sum"))
} else {
data <- data %>%
filter(year == yearOfInterest, country == countryName) %>%
select(-c(country, country_code, year))
}
data %>%
gather(key = "disease", value = "deaths") %>%
mutate(deaths = deaths / sum(deaths)) %>%
ggplot(aes(x = reorder(disease, deaths), y = deaths, fill = disease))+
geom_bar(stat = "identity")+
geom_text(aes(label = paste(round(100 * deaths, 2), "%")), hjust = -0.1)+
scale_y_continuous(labels = scales::percent, limits = c(0, 0.35))+
scale_fill_manual(values = colors)+
guides(fill = FALSE)+
coord_flip()+
xlab("")+
ylab("")+
theme_classic()+
labs(title = title, subtitle = subtitle, caption = caption)+
theme(
panel.grid.major.x = element_line(linetype = "dotted", color = "#5043484A")
)
}So let’s try it out on the dataset! First we import the data with readxl and use stringr to remove the percentage signs from the variable names. I also decided to omit NA’s, which I think is why my numbers are slightly different from those in the article. Oh well shrug
data <- readxl::read_xlsx("~/Code/tidy_tuesday/Week_3/global_mortality.xlsx") %>%
rename_all(funs(stringr::str_remove_all(., "[(%)]"))) %>%
na.omit()
data
## # A tibble: 4,524 x 35
## country country_code year `Cardiovascular… `Cancers ` `Respiratory di…
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Afghan… AFG 1990 17.6 4.03 2.11
## 2 Afghan… AFG 1991 17.8 4.05 2.13
## 3 Afghan… AFG 1992 18.4 4.17 2.21
## 4 Afghan… AFG 1994 19.1 4.26 2.31
## 5 Afghan… AFG 1995 19.9 4.39 2.41
## 6 Afghan… AFG 1996 20.4 4.48 2.49
## 7 Afghan… AFG 1997 20.4 4.45 2.49
## 8 Afghan… AFG 1998 19.6 4.26 2.41
## 9 Afghan… AFG 1999 21.1 4.57 2.60
## 10 Afghan… AFG 2000 21.4 4.64 2.64
## # … with 4,514 more rows, and 29 more variables: `Diabetes ` <dbl>,
## # `Dementia ` <dbl>, `Lower respiratory infections ` <dbl>, `Neonatal
## # deaths ` <dbl>, `Diarrheal diseases ` <dbl>, `Road accidents ` <dbl>,
## # `Liver disease ` <dbl>, `Tuberculosis ` <dbl>, `Kidney disease
## # ` <dbl>, `Digestive diseases ` <dbl>, `HIV/AIDS ` <dbl>, `Suicide
## # ` <dbl>, `Malaria ` <dbl>, `Homicide ` <dbl>, `Nutritional
## # deficiencies ` <dbl>, `Meningitis ` <dbl>, `Protein-energy
## # malnutrition ` <dbl>, `Drowning ` <dbl>, `Maternal deaths ` <dbl>,
## # `Parkinson disease ` <dbl>, `Alcohol disorders ` <dbl>, `Intestinal
## # infectious diseases ` <dbl>, `Drug disorders ` <dbl>, `Hepatitis
## # ` <dbl>, `Fire ` <dbl>, `Heat-related hot and cold exposure ` <dbl>,
## # `Natural disasters ` <dbl>, `Conflict ` <dbl>, `Terrorism ` <dbl>Finally, let’s use the function on the world, the US, and my beloved Chile for 2016.
plotting_function(data = data, yearOfInterest = 2016, countryName = "World")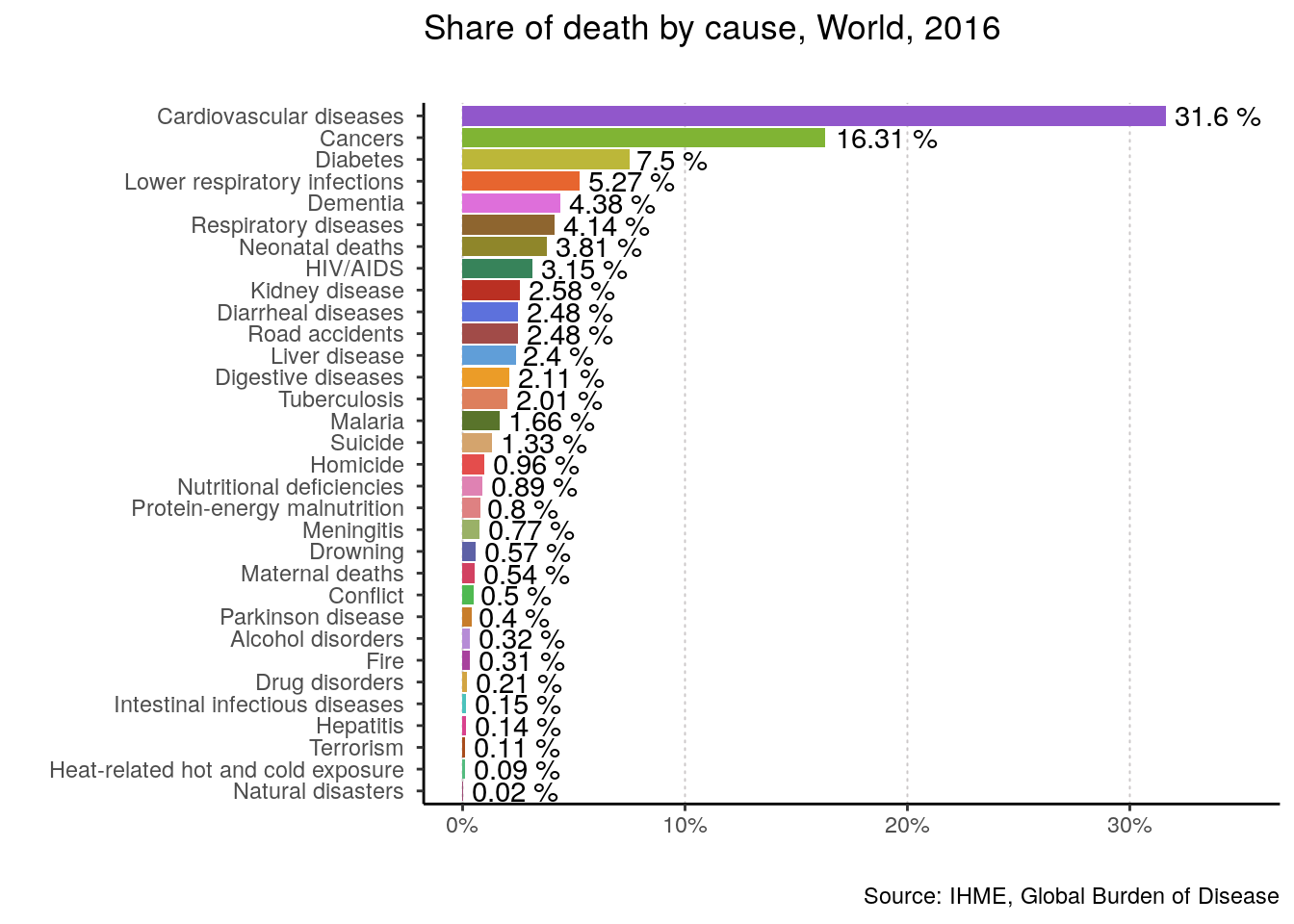
plotting_function(data = data, yearOfInterest = 2016, countryName = "United States")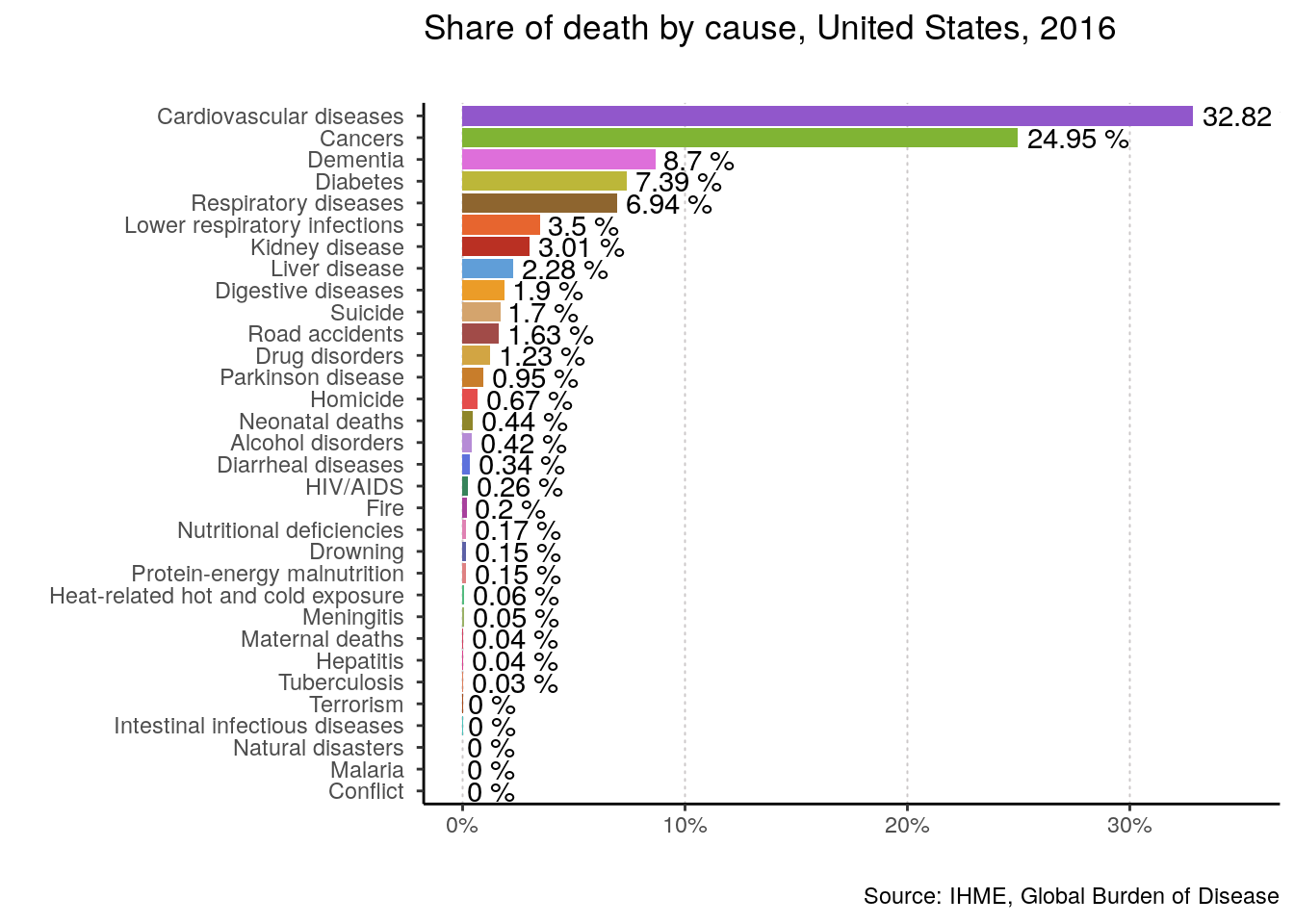
plotting_function(data = data, yearOfInterest = 2016, countryName = "Chile")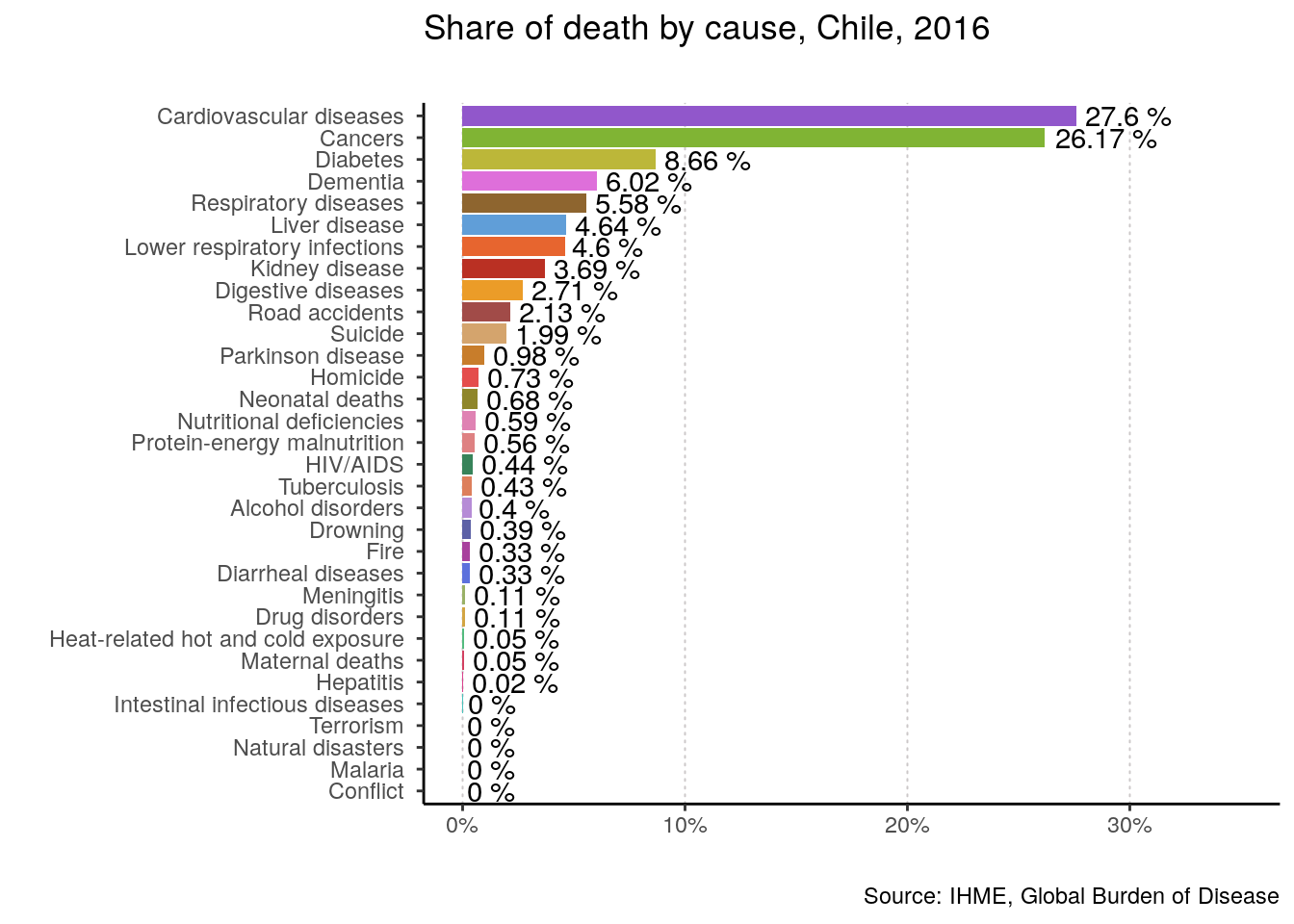
Nice! Of course, we could input any country and year and get the same kind of plot. That’s all friends!

Twitter
Google+
Facebook
Reddit
LinkedIn
StumbleUpon
Pinterest
Email